Atmospheric Retrievals with POSEIDON
At long last, your proposal to observe the newly discovered hot Jupiter WASP-999b with the Hubble Space Telescope has been accepted. Congratulations!
Loading Data
Months later, after carefully reducing the observations, you are ready to gaze in awe at your transmission spectrum.
First, you load all the usual stellar and planetary properties for this system.
[1]:
from POSEIDON.core import create_star, create_planet
from POSEIDON.constants import R_Sun, R_J
#***** Define stellar properties *****#
R_s = 1.155*R_Sun # Stellar radius (m)
T_s = 6071.0 # Stellar effective temperature (K)
Met_s = 0.0 # Stellar metallicity [log10(Fe/H_star / Fe/H_solar)]
log_g_s = 4.38 # Stellar log surface gravity (log10(cm/s^2) by convention)
# Create the stellar object
star = create_star(R_s, T_s, log_g_s, Met_s)
#***** Define planet properties *****#
planet_name = 'WASP-999b' # Planet name used for plots, output files etc.
R_p = 1.359*R_J # Planetary radius (m)
g_p = 9.186 # Gravitational field of planet (m/s^2)
T_eq = 1400.0 # Equilibrium temperature (K)
# Create the planet object
planet = create_planet(planet_name, R_p, gravity = g_p, T_eq = T_eq)
Next, you plot your observed transmission spectrum of WASP-999b.
[2]:
from POSEIDON.core import load_data, wl_grid_constant_R
from POSEIDON.visuals import plot_data
#***** Model wavelength grid *****#
wl_min = 0.4 # Minimum wavelength (um)
wl_max = 1.8 # Maximum wavelength (um)
R = 4000 # Spectral resolution of grid
# We need to provide a model wavelength grid to initialise instrument properties
wl = wl_grid_constant_R(wl_min, wl_max, R)
#***** Specify data location and instruments *****#
data_dir = '../../../POSEIDON/reference_data/observations/WASP-999b' # Change this to where your data is stored
datasets = ['WASP-999b_WFC3_G141.dat'] # Found in reference_data/observations
instruments = ['WFC3_G141'] # Instruments corresponding to the data
# Load dataset, pre-load instrument PSF and transmission function
data = load_data(data_dir, datasets, instruments, wl)
# Plot our data
fig_data = plot_data(data, planet_name)
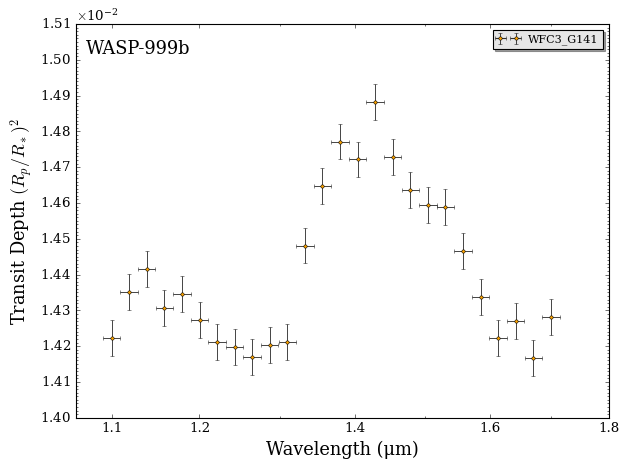
The spectrum isn’t flat! 🎉
Even better, your long term collaborator Dr. Tenalp just so happens to also have some observations of WASP-999b at shorter wavelengths. Let’s add their Hubble STIS data to our collection.
Note:
The data file format expected by POSEIDON is:
wavelength (μm) | bin half width (μm) | transit depth \((R_p/R_s)^2\) | transit depth error
[3]:
# Specify the STIS and WFC3 Hubble data
data_dir = '../../../POSEIDON/reference_data/observations/WASP-999b'
datasets = ['WASP-999b_STIS_G430.dat',
'WASP-999b_STIS_G750.dat',
'WASP-999b_WFC3_G141.dat']
instruments = ['STIS_G430', 'STIS_G750', 'WFC3_G141']
# Load dataset, pre-load instrument PSF and transmission function
data = load_data(data_dir, datasets, instruments, wl)
# Plot our data
fig_data = plot_data(data, planet_name)
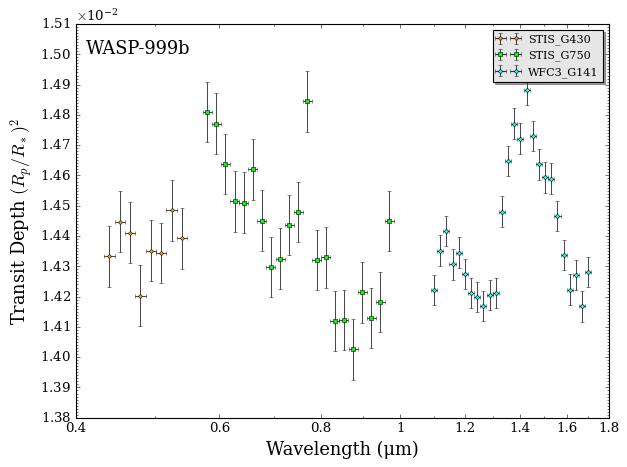
With data in hand, you are ready to begin the process of retrieving WASP-999b’s atmospheric properties.
Creating a Retrieval Model
Now comes the creative part: what model do you try first to fit WASP-999b’s transmission spectrum?
Given the a priori known low density of the planet, you conclude it is reasonable to assume this is a giant planet dominated by \(\rm{H}_2\) and \(\rm{He}\). Looking at your data above, especially the huge absorption feature in the infrared around 1.4 μm, you guess that \(\rm{H}_2 \rm{O}\) could be present (based on theoretical predictions or after looking up its cross section).
So for a first attempt, you consider a model with \(\rm{H}_2 \rm{O}\), an isothermal temperature profile, and no clouds.
[4]:
from POSEIDON.core import define_model
#***** Define model *****#
model_name = 'My_first_retrieval' # Model name used for plots, output files etc.
bulk_species = ['H2', 'He'] # H2 + He comprises the bulk atmosphere
param_species = ['H2O'] # The only trace gas is H2O
# Create the model object
model = define_model(model_name, bulk_species, param_species,
PT_profile = 'isotherm', cloud_model = 'cloud-free')
# Check the free parameters defining this model
print("Free parameters: " + str(model['param_names']))
Free parameters: ['R_p_ref' 'T' 'log_H2O']
Setting Retrieval Priors
One of the most important aspects in any Bayesian analysis is deciding what priors to use for the free parameters. Specifying a prior has two steps: (i) choosing the type of probability distribution; and (ii) choosing the allowable range.
Most free parameters in atmospheric retrievals with POSEIDON use the following prior types:
Uniform: you provide the minimum and maximum values for the parameter.
Gaussian: you provide the mean and standard deviation for the parameter.
Note:
If you do not specify a prior type or range for a given parameter, POSEIDON will ascribe a default prior type (generally uniform) and a ‘generous’ range.
Priors for WASP-999b
Your first retrieval is defined by three free parameters: (1) the isothermal atmospheric temperature; (2) the radius at the (fixed) reference pressure; and (3) the log-mixing ratio of \(\rm{H}_2 \rm{O}\). Since you don’t have any a priori information on WASP-999b’s atmosphere, you decide to use uniform priors for the three parameters.
You think a reasonable prior range for the temperature of this hot Jupiter is \(400 \, \rm{K}\) to \((T_{\rm{eq}} + 200 \, \rm{K}) = 1600 \, \rm{K}\). For the reference radius, you choose a wide range from 85% to 115% of the observed white light radius. Finally, for the \(\rm{H}_2 \rm{O}\) abundance you ascribe a very broad range from \(10^{-12}\) to 0.1.
[5]:
from POSEIDON.core import set_priors
#***** Set priors for retrieval *****#
# Initialise prior type dictionary
prior_types = {}
# Specify whether priors are linear, Gaussian, etc.
prior_types['T'] = 'uniform'
prior_types['R_p_ref'] = 'uniform'
prior_types['log_H2O'] = 'uniform'
# Initialise prior range dictionary
prior_ranges = {}
# Specify prior ranges for each free parameter
prior_ranges['T'] = [400, 1600]
prior_ranges['R_p_ref'] = [0.85*R_p, 1.15*R_p]
prior_ranges['log_H2O'] = [-12, -1]
# Create prior object for retrieval
priors = set_priors(planet, star, model, data, prior_types, prior_ranges)
Pre-load Opacities
The last step before running a retrieval is to pre-interpolate the cross sections for our model and store them in memory. For more details on this process, see the forward model tutorial.
Warning:
Ensure the range of \(T_{\rm{fine}}\) used for opacity pre-interpolation is at least as large as the desired prior range for temperatures to be explored in the retrieval. Any models with layer temperatures falling outside the range of \(T_{\rm{fine}}\) will be automatically rejected (for retrievals with non-isothermal P-T profiles, this prevents unphysical profiles with negative temperatures etc.)
[6]:
from POSEIDON.core import read_opacities
import numpy as np
#***** Read opacity data *****#
opacity_treatment = 'opacity_sampling'
# Define fine temperature grid (K)
T_fine_min = 400 # Same as prior range for T
T_fine_max = 1600 # Same as prior range for T
T_fine_step = 10 # 10 K steps are a good tradeoff between accuracy and RAM
T_fine = np.arange(T_fine_min, (T_fine_max + T_fine_step), T_fine_step)
# Define fine pressure grid (log10(P/bar))
log_P_fine_min = -6.0 # 1 ubar is the lowest pressure in the opacity database
log_P_fine_max = 2.0 # 100 bar is the highest pressure in the opacity database
log_P_fine_step = 0.2 # 0.2 dex steps are a good tradeoff between accuracy and RAM
log_P_fine = np.arange(log_P_fine_min, (log_P_fine_max + log_P_fine_step),
log_P_fine_step)
# Pre-interpolate the opacities
opac = read_opacities(model, wl, opacity_treatment, T_fine, log_P_fine)
Reading in cross sections in opacity sampling mode...
H2-H2 done
H2-He done
H2O done
Opacity pre-interpolation complete.
Run Retrieval
You are now ready to run your first atmospheric retrieval!
Here we will use the nested sampling algorithm MultiNest to explore the parameter space. The key input quantity you need to provide to MultiNest is called the number of live points, \(N_{\rm{live}}\), which determines how finely the parameter space will be sampled (and hence the number of computed spectra). For exploratory retrievals, \(N_{\rm{live}} = 400\) usually suffices. For publication-quality results, \(N_{\rm{live}} = 2000\) is reasonable.
This simple POSEIDON retrieval should take about 10 minutes on 1 core for a typical laptop.
Tip:
Retrievals run faster on multiple cores. When running the cells in this Jupyter notebook, only a single core will be used. You can run a multi-core retrieval on 4 cores by converting this Jupyter notebook into a python script, then calling mpirun on the .py file:
mpirun -n 4 python -u YOUR_RETRIEVAL_SCRIPT.py
[7]:
from POSEIDON.retrieval import run_retrieval
#***** Specify fixed atmospheric settings for retrieval *****#
# Atmospheric pressure grid
P_min = 1.0e-7 # 0.1 ubar
P_max = 100 # 100 bar
N_layers = 100 # 100 layers
# Let's space the layers uniformly in log-pressure
P = np.logspace(np.log10(P_max), np.log10(P_min), N_layers)
# Specify the reference pressure
P_ref = 10.0 # Retrieved R_p_ref parameter will be the radius at 10 bar
#***** Run atmospheric retrieval *****#
run_retrieval(planet, star, model, opac, data, priors, wl, P, P_ref, R = R,
spectrum_type = 'transmission', sampling_algorithm = 'MultiNest',
N_live = 400, verbose = True)
POSEIDON now running 'My_first_retrieval'
*****************************************************
MultiNest v3.10
Copyright Farhan Feroz & Mike Hobson
Release Jul 2015
no. of live points = 400
dimensionality = 3
*****************************************************
Starting MultiNest
generating live points
live points generated, starting sampling
Acceptance Rate: 0.991189
Replacements: 450
Total Samples: 454
Nested Sampling ln(Z): -149376.393187
Acceptance Rate: 0.967118
Replacements: 500
Total Samples: 517
Nested Sampling ln(Z): -100223.232426
Acceptance Rate: 0.941781
Replacements: 550
Total Samples: 584
Nested Sampling ln(Z): -75907.575465
Acceptance Rate: 0.911854
Replacements: 600
Total Samples: 658
Nested Sampling ln(Z): -61093.857681
Acceptance Rate: 0.895317
Replacements: 650
Total Samples: 726
Nested Sampling ln(Z): -46918.636301
Acceptance Rate: 0.876095
Replacements: 700
Total Samples: 799
Nested Sampling ln(Z): -34709.259713
Acceptance Rate: 0.848416
Replacements: 750
Total Samples: 884
Nested Sampling ln(Z): -27917.745250
Acceptance Rate: 0.821355
Replacements: 800
Total Samples: 974
Nested Sampling ln(Z): -20817.638435
Acceptance Rate: 0.801131
Replacements: 850
Total Samples: 1061
Nested Sampling ln(Z): -16602.025529
Acceptance Rate: 0.787402
Replacements: 900
Total Samples: 1143
Nested Sampling ln(Z): -12828.795280
Acceptance Rate: 0.781250
Replacements: 950
Total Samples: 1216
Nested Sampling ln(Z): -9154.875724
Acceptance Rate: 0.767460
Replacements: 1000
Total Samples: 1303
Nested Sampling ln(Z): -6618.828263
Acceptance Rate: 0.761421
Replacements: 1050
Total Samples: 1379
Nested Sampling ln(Z): -5184.166098
Acceptance Rate: 0.763359
Replacements: 1100
Total Samples: 1441
Nested Sampling ln(Z): -3726.955025
Acceptance Rate: 0.759577
Replacements: 1150
Total Samples: 1514
Nested Sampling ln(Z): -3059.929611
Acceptance Rate: 0.756144
Replacements: 1200
Total Samples: 1587
Nested Sampling ln(Z): -2107.612314
Acceptance Rate: 0.752106
Replacements: 1250
Total Samples: 1662
Nested Sampling ln(Z): -1462.679418
Acceptance Rate: 0.748848
Replacements: 1300
Total Samples: 1736
Nested Sampling ln(Z): -1062.417515
Acceptance Rate: 0.744212
Replacements: 1350
Total Samples: 1814
Nested Sampling ln(Z): -780.193255
Acceptance Rate: 0.732218
Replacements: 1400
Total Samples: 1912
Nested Sampling ln(Z): -540.726532
Acceptance Rate: 0.727546
Replacements: 1450
Total Samples: 1993
Nested Sampling ln(Z): -373.767064
Acceptance Rate: 0.721154
Replacements: 1500
Total Samples: 2080
Nested Sampling ln(Z): -264.632893
Acceptance Rate: 0.717593
Replacements: 1550
Total Samples: 2160
Nested Sampling ln(Z): -181.260821
Acceptance Rate: 0.714286
Replacements: 1600
Total Samples: 2240
Nested Sampling ln(Z): -107.919284
Acceptance Rate: 0.706941
Replacements: 1650
Total Samples: 2334
Nested Sampling ln(Z): -53.221660
Acceptance Rate: 0.701899
Replacements: 1700
Total Samples: 2422
Nested Sampling ln(Z): -15.078571
Acceptance Rate: 0.700000
Replacements: 1750
Total Samples: 2500
Nested Sampling ln(Z): 19.087249
Acceptance Rate: 0.697134
Replacements: 1800
Total Samples: 2582
Nested Sampling ln(Z): 49.342309
Acceptance Rate: 0.697587
Replacements: 1850
Total Samples: 2652
Nested Sampling ln(Z): 72.205019
Acceptance Rate: 0.694698
Replacements: 1900
Total Samples: 2735
Nested Sampling ln(Z): 85.943300
Acceptance Rate: 0.692718
Replacements: 1950
Total Samples: 2815
Nested Sampling ln(Z): 97.244655
Acceptance Rate: 0.691563
Replacements: 2000
Total Samples: 2892
Nested Sampling ln(Z): 104.984281
Acceptance Rate: 0.691633
Replacements: 2050
Total Samples: 2964
Nested Sampling ln(Z): 113.403592
Acceptance Rate: 0.688299
Replacements: 2100
Total Samples: 3051
Nested Sampling ln(Z): 131.764879
Acceptance Rate: 0.686024
Replacements: 2150
Total Samples: 3134
Nested Sampling ln(Z): 157.360852
Acceptance Rate: 0.685999
Replacements: 2200
Total Samples: 3207
Nested Sampling ln(Z): 180.557511
Acceptance Rate: 0.686604
Replacements: 2250
Total Samples: 3277
Nested Sampling ln(Z): 196.282332
Acceptance Rate: 0.682088
Replacements: 2300
Total Samples: 3372
Nested Sampling ln(Z): 216.905503
Acceptance Rate: 0.677233
Replacements: 2350
Total Samples: 3470
Nested Sampling ln(Z): 232.444613
Acceptance Rate: 0.674157
Replacements: 2400
Total Samples: 3560
Nested Sampling ln(Z): 245.752503
Acceptance Rate: 0.673447
Replacements: 2450
Total Samples: 3638
Nested Sampling ln(Z): 257.105847
Acceptance Rate: 0.672405
Replacements: 2500
Total Samples: 3718
Nested Sampling ln(Z): 264.317581
Acceptance Rate: 0.671053
Replacements: 2550
Total Samples: 3800
Nested Sampling ln(Z): 272.399048
Acceptance Rate: 0.670276
Replacements: 2600
Total Samples: 3879
Nested Sampling ln(Z): 280.718449
Acceptance Rate: 0.669361
Replacements: 2650
Total Samples: 3959
Nested Sampling ln(Z): 287.320478
Acceptance Rate: 0.667326
Replacements: 2700
Total Samples: 4046
Nested Sampling ln(Z): 293.412676
Acceptance Rate: 0.668287
Replacements: 2750
Total Samples: 4115
Nested Sampling ln(Z): 298.766003
Acceptance Rate: 0.666032
Replacements: 2800
Total Samples: 4204
Nested Sampling ln(Z): 304.690107
Acceptance Rate: 0.660487
Replacements: 2850
Total Samples: 4315
Nested Sampling ln(Z): 309.835432
Acceptance Rate: 0.658492
Replacements: 2900
Total Samples: 4404
Nested Sampling ln(Z): 314.350706
Acceptance Rate: 0.658923
Replacements: 2950
Total Samples: 4477
Nested Sampling ln(Z): 318.889920
Acceptance Rate: 0.657318
Replacements: 3000
Total Samples: 4564
Nested Sampling ln(Z): 322.338650
Acceptance Rate: 0.656055
Replacements: 3050
Total Samples: 4649
Nested Sampling ln(Z): 325.644907
Acceptance Rate: 0.654976
Replacements: 3100
Total Samples: 4733
Nested Sampling ln(Z): 329.504780
Acceptance Rate: 0.650961
Replacements: 3150
Total Samples: 4839
Nested Sampling ln(Z): 332.967266
Acceptance Rate: 0.646987
Replacements: 3200
Total Samples: 4946
Nested Sampling ln(Z): 335.740677
Acceptance Rate: 0.645225
Replacements: 3250
Total Samples: 5037
Nested Sampling ln(Z): 337.992870
Acceptance Rate: 0.644405
Replacements: 3300
Total Samples: 5121
Nested Sampling ln(Z): 340.644460
Acceptance Rate: 0.641885
Replacements: 3350
Total Samples: 5219
Nested Sampling ln(Z): 343.085635
Acceptance Rate: 0.638618
Replacements: 3400
Total Samples: 5324
Nested Sampling ln(Z): 345.024246
Acceptance Rate: 0.637708
Replacements: 3450
Total Samples: 5410
Nested Sampling ln(Z): 346.667340
Acceptance Rate: 0.636943
Replacements: 3500
Total Samples: 5495
Nested Sampling ln(Z): 348.223073
Acceptance Rate: 0.636429
Replacements: 3550
Total Samples: 5578
Nested Sampling ln(Z): 349.879637
Acceptance Rate: 0.636830
Replacements: 3600
Total Samples: 5653
Nested Sampling ln(Z): 351.323517
Acceptance Rate: 0.635999
Replacements: 3650
Total Samples: 5739
Nested Sampling ln(Z): 352.676886
Acceptance Rate: 0.633453
Replacements: 3700
Total Samples: 5841
Nested Sampling ln(Z): 353.976060
Acceptance Rate: 0.632484
Replacements: 3750
Total Samples: 5929
Nested Sampling ln(Z): 355.110099
Acceptance Rate: 0.631544
Replacements: 3800
Total Samples: 6017
Nested Sampling ln(Z): 356.111924
Acceptance Rate: 0.631148
Replacements: 3850
Total Samples: 6100
Nested Sampling ln(Z): 356.954789
Acceptance Rate: 0.631988
Replacements: 3900
Total Samples: 6171
Nested Sampling ln(Z): 357.659505
Acceptance Rate: 0.632202
Replacements: 3950
Total Samples: 6248
Nested Sampling ln(Z): 358.314164
Acceptance Rate: 0.631612
Replacements: 4000
Total Samples: 6333
Nested Sampling ln(Z): 358.986584
Acceptance Rate: 0.630546
Replacements: 4050
Total Samples: 6423
Nested Sampling ln(Z): 359.655445
Acceptance Rate: 0.629220
Replacements: 4100
Total Samples: 6516
Nested Sampling ln(Z): 360.260259
Acceptance Rate: 0.629169
Replacements: 4150
Total Samples: 6596
Nested Sampling ln(Z): 360.790076
Acceptance Rate: 0.629025
Replacements: 4200
Total Samples: 6677
Nested Sampling ln(Z): 361.301084
Acceptance Rate: 0.628234
Replacements: 4250
Total Samples: 6765
Nested Sampling ln(Z): 361.766874
Acceptance Rate: 0.628655
Replacements: 4300
Total Samples: 6840
Nested Sampling ln(Z): 362.155539
Acceptance Rate: 0.629431
Replacements: 4350
Total Samples: 6911
Nested Sampling ln(Z): 362.545721
Acceptance Rate: 0.630192
Replacements: 4400
Total Samples: 6982
Nested Sampling ln(Z): 362.913660
Acceptance Rate: 0.629064
Replacements: 4450
Total Samples: 7074
Nested Sampling ln(Z): 363.250782
Acceptance Rate: 0.628755
Replacements: 4500
Total Samples: 7157
Nested Sampling ln(Z): 363.540926
Acceptance Rate: 0.628366
Replacements: 4550
Total Samples: 7241
Nested Sampling ln(Z): 363.791715
Acceptance Rate: 0.628415
Replacements: 4600
Total Samples: 7320
Nested Sampling ln(Z): 364.027008
Acceptance Rate: 0.628293
Replacements: 4650
Total Samples: 7401
Nested Sampling ln(Z): 364.239225
Acceptance Rate: 0.628594
Replacements: 4700
Total Samples: 7477
Nested Sampling ln(Z): 364.438803
Acceptance Rate: 0.628224
Replacements: 4750
Total Samples: 7561
Nested Sampling ln(Z): 364.626559
Acceptance Rate: 0.628272
Replacements: 4800
Total Samples: 7640
Nested Sampling ln(Z): 364.793035
Acceptance Rate: 0.628727
Replacements: 4850
Total Samples: 7714
Nested Sampling ln(Z): 364.941865
Acceptance Rate: 0.628689
Replacements: 4900
Total Samples: 7794
Nested Sampling ln(Z): 365.073906
Acceptance Rate: 0.628731
Replacements: 4950
Total Samples: 7873
Nested Sampling ln(Z): 365.197704
Acceptance Rate: 0.628773
Replacements: 5000
Total Samples: 7952
Nested Sampling ln(Z): 365.309154
Acceptance Rate: 0.628422
Replacements: 5050
Total Samples: 8036
Nested Sampling ln(Z): 365.411631
Acceptance Rate: 0.628156
Replacements: 5100
Total Samples: 8119
Nested Sampling ln(Z): 365.504425
Acceptance Rate: 0.628125
Replacements: 5150
Total Samples: 8199
Nested Sampling ln(Z): 365.587869
Acceptance Rate: 0.628247
Replacements: 5200
Total Samples: 8277
Nested Sampling ln(Z): 365.662347
Acceptance Rate: 0.628968
Replacements: 5250
Total Samples: 8347
Nested Sampling ln(Z): 365.729805
Acceptance Rate: 0.627293
Replacements: 5300
Total Samples: 8449
Nested Sampling ln(Z): 365.790244
Acceptance Rate: 0.626684
Replacements: 5350
Total Samples: 8537
Nested Sampling ln(Z): 365.844231
Acceptance Rate: 0.625145
Replacements: 5400
Total Samples: 8638
Nested Sampling ln(Z): 365.892719
Acceptance Rate: 0.626005
Replacements: 5450
Total Samples: 8706
Nested Sampling ln(Z): 365.936024
Acceptance Rate: 0.625355
Replacements: 5500
Total Samples: 8795
Nested Sampling ln(Z): 365.975268
Acceptance Rate: 0.625070
Replacements: 5550
Total Samples: 8879
Nested Sampling ln(Z): 366.010460
Acceptance Rate: 0.624761
Replacements: 5556
Total Samples: 8893
Nested Sampling ln(Z): 366.014423
POSEIDON retrieval finished in 0.05 hours
ln(ev)= 366.30326117403126 +/- 0.16121614119644562
Total Likelihood Evaluations: 8893
Sampling finished. Exiting MultiNest
Now generating 1000 sampled spectra and P-T profiles from the posterior distribution...
This process will take approximately 0.33 minutes
All done! Output files can be found in ./POSEIDON_output/WASP-999b/retrievals/results/
Plot Retrieval Results
Now that the retrieval is finished, you’re eager and ready to see what WASP-999b’s atmosphere is hiding.
You first plot confidence intervals of the retrieved spectrum from this model compared to WASP-999b’s observed transmission spectrum. You also generate a corner plot showing the retrieved probability distributions of the model parameters.
[9]:
from POSEIDON.utility import read_retrieved_spectrum, plot_collection
from POSEIDON.visuals import plot_spectra_retrieved
from POSEIDON.corner import generate_cornerplot
#***** Plot retrieved transmission spectrum *****#
# Read retrieved spectrum confidence regions
wl, spec_low2, spec_low1, spec_median, \
spec_high1, spec_high2 = read_retrieved_spectrum(planet_name, model_name)
# Create composite spectra objects for plotting
spectra_median = plot_collection(spec_median, wl, collection = [])
spectra_low1 = plot_collection(spec_low1, wl, collection = [])
spectra_low2 = plot_collection(spec_low2, wl, collection = [])
spectra_high1 = plot_collection(spec_high1, wl, collection = [])
spectra_high2 = plot_collection(spec_high2, wl, collection = [])
# Produce figure
fig_spec = plot_spectra_retrieved(spectra_median, spectra_low2, spectra_low1,
spectra_high1, spectra_high2, planet_name,
data, R_to_bin = 100,
data_labels = ['STIS G430', 'STIS G750', 'WFC3 G141'],
data_colour_list = ['lime', 'cyan', 'black'])
#***** Make corner plot *****#
fig_corner = generate_cornerplot(planet, model)
Generating corner plot ...
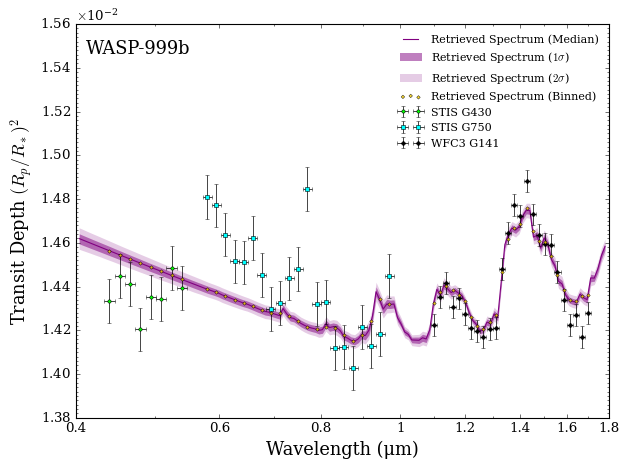
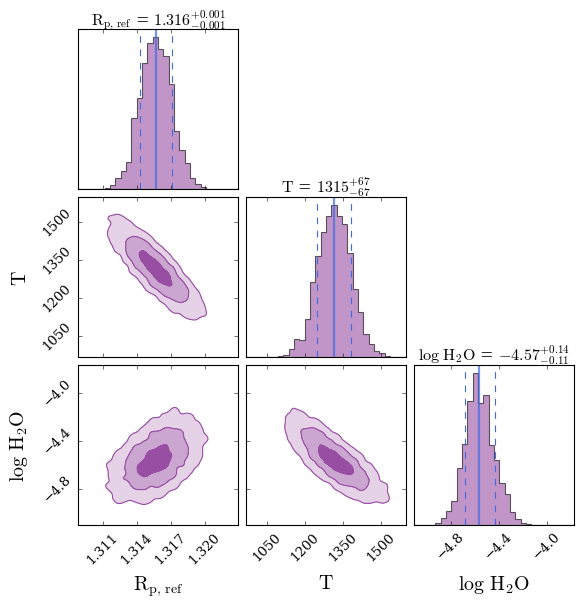
Not bad for a first simple model! The fit to the infrared WFC3 data looks reasonable, but there is considerable scatter in the visible wavelength data which isn’t captured by the model. You quantitatively assess the fit quality by opening the retrieval results file.
Tip:
Retrieval results are automatically created in the POSEIDON output directory, which will appear in the same directory as the Python file running POSEIDON.
The main retrieval results are found in
./POSEIDON_output/𝗽𝗹𝗮𝗻𝗲𝘁_𝗻𝗮𝗺𝗲/retrievals/results
There, you will find 2 files for each retrieval:
𝗺𝗼𝗱𝗲𝗹_𝗻𝗮𝗺𝗲_corner.pdf — the corner plot.
𝗺𝗼𝗱𝗲𝗹_𝗻𝗮𝗺𝗲_results.txt — a human-readable summary of the main retrieval results.
You can also find the posterior samples from the retrieval in
./POSEIDON_output/𝗽𝗹𝗮𝗻𝗲𝘁_𝗻𝗮𝗺𝗲/retrievals/samples
Inside the results file, you scroll down to the fit quality statistics.

Well, that reduced \(\chi^2\) doesn’t look great… a value significantly greater than 1 likely indicates that your model is under fitting the observations.
Comparing Retrieval Fits
After a little reflection, you suspect the issue is that one or more chemical species with strong absorption cross sections at visible wavelengths is missing from the model. So you define a new model with \(\rm{Na}\), \(\rm{K}\), and \(\rm{TiO}\) added and run a second retrieval.
This 6-parameter retrieval should take about 20 minutes on a single core.
[9]:
#***** Define new model *****#
model_name_2 = 'Improved_retrieval'
bulk_species = ['H2', 'He']
param_species_2 = ['Na', 'K', 'TiO', 'H2O'] # Three new chemical species added
# Create the model object
model_2 = define_model(model_name_2, bulk_species, param_species_2,
PT_profile = 'isotherm', cloud_model = 'cloud-free')
# Check the free parameters defining this model
print("Free parameters: " + str(model_2['param_names']))
#***** Set priors for new retrieval *****#
# Initialise prior type dictionary
prior_types_2 = {}
# Specify whether priors are linear, Gaussian, etc.
prior_types_2['T'] = 'uniform'
prior_types_2['R_p_ref'] = 'uniform'
prior_types_2['log_X'] = 'uniform' # 'log_X' sets the same prior for all mixing ratios
# Initialise prior range dictionary
prior_ranges_2 = {}
# Specify prior ranges for each free parameter
prior_ranges_2['T'] = [400, 1600]
prior_ranges_2['R_p_ref'] = [0.85*R_p, 1.15*R_p]
prior_ranges_2['log_X'] = [-12, -1] # 'log_X' sets the same prior for all mixing ratios
# Create prior object for retrieval
priors_2 = set_priors(planet, star, model_2, data, prior_types_2, prior_ranges_2)
#***** Read opacity data *****#
# Pre-interpolate the opacities
opac_2 = read_opacities(model_2, wl, opacity_treatment, T_fine, log_P_fine)
#***** Run atmospheric retrieval *****#
run_retrieval(planet, star, model_2, opac_2, data, priors_2, wl, P, P_ref, R = R,
spectrum_type = 'transmission', sampling_algorithm = 'MultiNest',
N_live = 400, verbose = False) # This last variable suppresses MultiNest terminal output
Free parameters: ['R_p_ref' 'T' 'log_Na' 'log_K' 'log_TiO' 'log_H2O']
Reading in cross sections in opacity sampling mode...
H2-H2 done
H2-He done
Na done
K done
TiO done
H2O done
Opacity pre-interpolation complete.
POSEIDON now running 'Improved_retrieval'
*****************************************************
MultiNest v3.10
Copyright Farhan Feroz & Mike Hobson
Release Jul 2015
no. of live points = 400
dimensionality = 6
*****************************************************
POSEIDON retrieval finished in 0.15 hours
ln(ev)= 433.18950611189564 +/- 0.19726984263604322
Total Likelihood Evaluations: 22727
Sampling finished. Exiting MultiNest
Now generating 1000 sampled spectra and P-T profiles from the posterior distribution...
This process will take approximately 0.39 minutes
All done! Output files can be found in ./POSEIDON_output/WASP-999b/retrievals/results/
Now that the new retrieval has finished, you compare the fit quality between the two models.
[10]:
#***** Plot retrieved transmission spectrum *****#
# Read retrieved spectrum confidence regions
wl, spec_low2, spec_low1, spec_median, \
spec_high1, spec_high2 = read_retrieved_spectrum(planet_name, model_name_2)
# Create composite spectra objects for plotting
spectra_median = plot_collection(spec_median, wl, collection = spectra_median)
spectra_low1 = plot_collection(spec_low1, wl, collection = spectra_low1)
spectra_low2 = plot_collection(spec_low2, wl, collection = spectra_low2)
spectra_high1 = plot_collection(spec_high1, wl, collection = spectra_high1)
spectra_high2 = plot_collection(spec_high2, wl, collection = spectra_high2)
# Produce figure
fig_spec = plot_spectra_retrieved(spectra_median, spectra_low2, spectra_low1,
spectra_high1, spectra_high2, planet_name,
data, R_to_bin = 100,
spectra_labels = ['First Model', 'Add Alkalis'],
data_labels = ['STIS G430', 'STIS G750', 'WFC3 G141'],
data_colour_list = ['lime', 'cyan', 'black'])
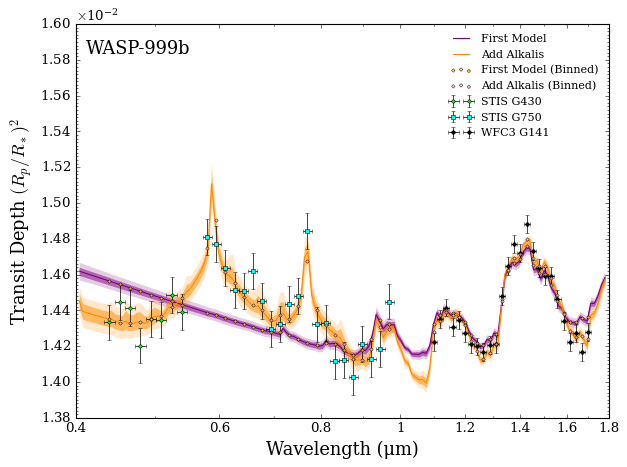
Now that looks like a much better fit! Checking the results file, you see that the Bayesian evidence for this new model is significantly higher and the reduced \(\chi^2\) is now much closer to 1.

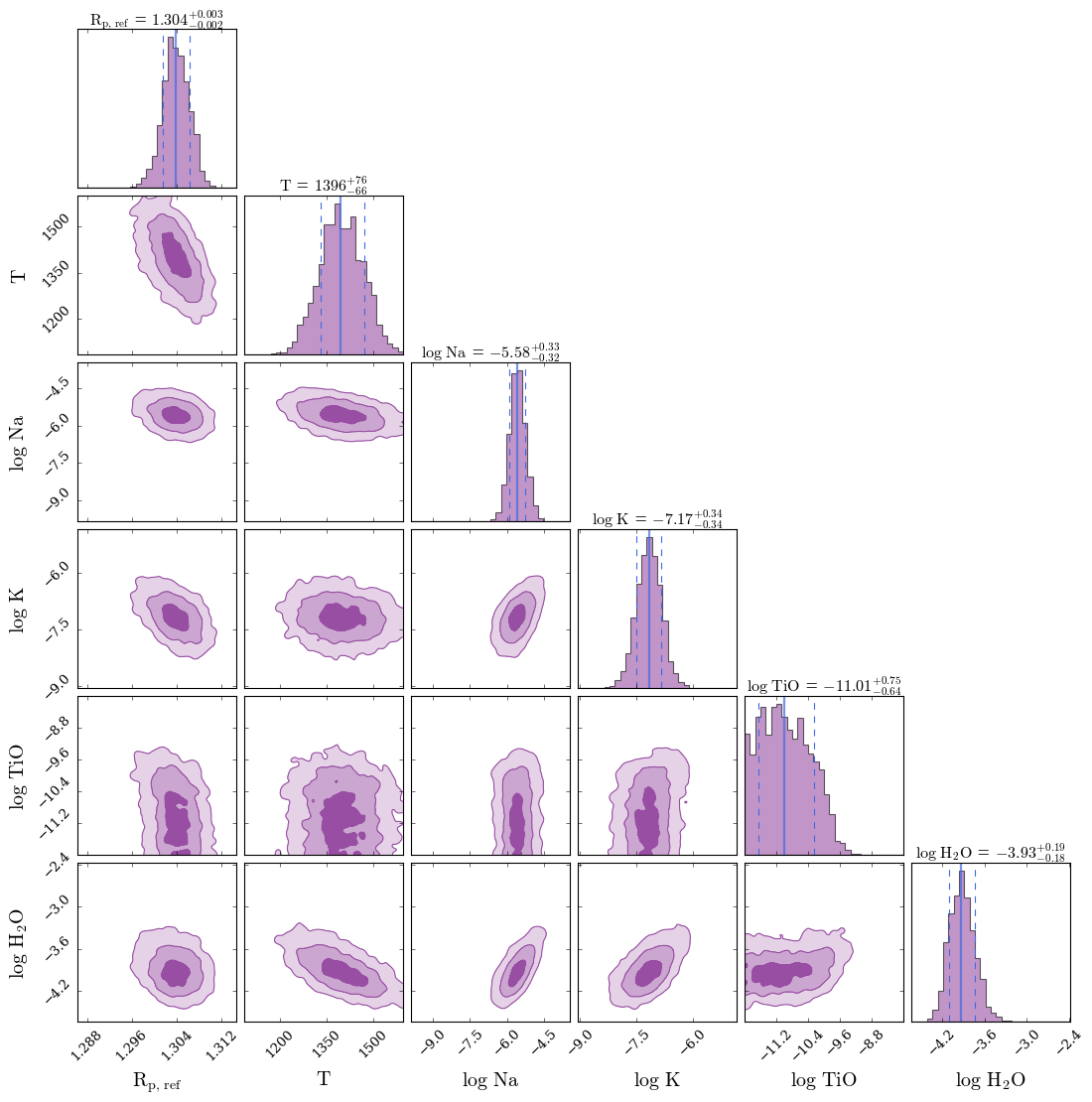
In fact, given that the reduced \(\chi^2 < 1\), this suggests a degree of over fitting. To see what is going on, you create a corner plot to visualise the retrieved parameters.
[11]:
fig_corner = generate_cornerplot(planet, model_2)
Generating corner plot ...
Bayesian Model Comparisons
What we have seen above are examples of Bayesian parameter estimation, represented by the corner plots (or more formally, the joint posterior probability distribution). Parameter estimation problems ask the question, “Given a model, what are the ranges of the model parameters that fit this dataset?”.
But we can also pose the question, “Given many models, which are better at explaining the data?”. This type of analysis is known as Bayesian model comparison.
Model Comparison Statistics
You already saw an example of a simple model comparison metric above (the reduced \(\chi^2\) of the best-fitting parameter combination), but we can go further. One of the by-products of a POSEIDON retrieval with MultiNest is the Bayesian evidence, \(\mathcal{Z}\), which is a global metric quantifying the quality of the model fit to the data (mathematically, the normalisation factor in the denominator of Bayes theorem). You actually saw the evidence values already in the screenshots from the POSEIDON ‘results.txt’ files.
While the Bayesian evidence of a single model isn’t informative, the ratio of the evidences of two nested models, the Bayes factor, \(\mathcal{B_{12}}\), tells us the probability (odds) ratio for the first model compared to the second model.
In the atmospheric retrieval literature, it is also common to see ‘frequentist equivalent’ detection significances quotes (expressed as a number of \(\sigma\)).
Chemical Detection Significances
Let’s now apply Bayesian model comparisons to answer the following question:
How confident are we that \(\rm{H}_2 \rm{O}\) and \(\rm{TiO}\) are in WASP-999b’s atmosphere?
To answer this, we first need to chose a reference model. Since our improved model above (with \(\rm{Na}\), \(\rm{K}\), \(\rm{TiO}\), and \(\rm{H}_2 \rm{O}\)) provides a good fit to the observations, we will adopt it as our reference model.
We will now run two more retrievals, both with the same setup as reference model but with (i) \(\rm{H}_2 \rm{O}\) removed and (ii) \(\rm{TiO}\) removed. These models are therefore nested models with respect to our reference model, since they have one less free parameter.
[12]:
#***** Define nested models *****#
model_name_3 = 'No_H2O'
model_name_4 = 'No_TiO'
param_species_3 = ['Na', 'K', 'TiO'] # No H2O
param_species_4 = ['Na', 'K', 'H2O'] # No TiO
# Create the model objects
model_3 = define_model(model_name_3, bulk_species, param_species_3,
PT_profile = 'isotherm', cloud_model = 'cloud-free')
model_4 = define_model(model_name_4, bulk_species, param_species_4,
PT_profile = 'isotherm', cloud_model = 'cloud-free')
# We can use the same priors as model 2 (since we used the same prior for all mixing ratios)
#***** Read opacity data *****#
# Pre-interpolate the opacities
opac_3 = read_opacities(model_3, wl, opacity_treatment, T_fine, log_P_fine)
opac_4 = read_opacities(model_4, wl, opacity_treatment, T_fine, log_P_fine)
#***** Run atmospheric retrievals *****#
# Retrieval with no H2O
run_retrieval(planet, star, model_3, opac_3, data, priors_2, wl, P, P_ref, R = R, # Same priors as the reference model
spectrum_type = 'transmission', sampling_algorithm = 'MultiNest',
N_live = 400, verbose = False)
# Retrieval with no TiO
run_retrieval(planet, star, model_4, opac_4, data, priors_2, wl, P, P_ref, R = R, # Same priors as the reference model
spectrum_type = 'transmission', sampling_algorithm = 'MultiNest',
N_live = 400, verbose = False)
Reading in cross sections in opacity sampling mode...
H2-H2 done
H2-He done
Na done
K done
TiO done
Opacity pre-interpolation complete.
Reading in cross sections in opacity sampling mode...
H2-H2 done
H2-He done
Na done
K done
H2O done
Opacity pre-interpolation complete.
POSEIDON now running 'No_H2O'
*****************************************************
MultiNest v3.10
Copyright Farhan Feroz & Mike Hobson
Release Jul 2015
no. of live points = 400
dimensionality = 5
*****************************************************
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
MultiNest Warning!
Parameter 2 of mode 1 is converging towards the edge of the prior.
POSEIDON retrieval finished in 0.089 hours
ln(ev)= 161.42808609101985 +/- 0.19555894644922608
Total Likelihood Evaluations: 14037
Sampling finished. Exiting MultiNest
Now generating 1000 sampled spectra and P-T profiles from the posterior distribution...
This process will take approximately 0.37 minutes
All done! Output files can be found in ./POSEIDON_output/WASP-999b/retrievals/results/
POSEIDON now running 'No_TiO'
*****************************************************
MultiNest v3.10
Copyright Farhan Feroz & Mike Hobson
Release Jul 2015
no. of live points = 400
dimensionality = 5
*****************************************************
POSEIDON retrieval finished in 0.11 hours
ln(ev)= 434.51587474756087 +/- 0.19056676348009649
Total Likelihood Evaluations: 17635
Sampling finished. Exiting MultiNest
Now generating 1000 sampled spectra and P-T profiles from the posterior distribution...
This process will take approximately 0.39 minutes
All done! Output files can be found in ./POSEIDON_output/WASP-999b/retrievals/results/
(By the way, when MultiNest prints out convergence warnings like those above it is a clue telling you the fit is so bad that another free parameter is being forced to the edge of the prior — in this case, it is because \(\rm{H}_2 \rm{O}\) is the only infrared opacity source so it really needs to be included in the model!)
POSEIDON includes a convenient function we can use to carry out Bayesian model comparisons.
[6]:
from POSEIDON.retrieval import Bayesian_model_comparison
# Rename the models just for convenience
model_ref = model_2
model_no_H2O = model_3
model_no_TiO = model_4
# H2O model comparison
Bayesian_model_comparison(planet_name, model_ref, model_no_H2O)
Bayesian evidences:
Model Improved_retrieval: ln Z = 433.19 +/- 0.20
Model No_H2O: ln Z = 161.43 +/- 0.20
Bayes factor:
B = 1.06e+118
ln B = 271.76
'Equivalent' detection significance:
23.5 σ
[7]:
# TiO model comparison
Bayesian_model_comparison(planet_name, model_ref, model_no_TiO)
Bayesian evidences:
Model Improved_retrieval: ln Z = 433.19 +/- 0.20
Model No_TiO: ln Z = 434.52 +/- 0.19
Bayes factor:
B = 2.65e-01
ln B = -1.33
No detection of the reference model, model No_TiO is preferred.
So the Bayesian model comparisons tell us we have a strong detection of \(\rm{H}_2 \rm{O}\) but no evidence for \(\rm{TiO}\). Indeed, the Bayesian evidence actually improves when we remove \(\rm{TiO}\).
The tendency of the Bayesian evidence to lower when including redundant parameters, sometimes called the ‘Occam penalty’, is one of the powerful features of the Bayesian evidence in finding the optimal model to explain a given dataset.
To gain intuition for why the model comparison so strongly favours \(\rm{H}_2 \rm{O}\), let’s plot the retrieved spectra.
[10]:
# Read retrieved spectrum confidence regions
wl, spec_low2, spec_low1, spec_median, \
spec_high1, spec_high2 = read_retrieved_spectrum(planet_name, model_ref['model_name'])
# Create composite spectra objects for plotting
spectra_median = plot_collection(spec_median, wl, collection = [])
spectra_low1 = plot_collection(spec_low1, wl, collection = [])
spectra_low2 = plot_collection(spec_low2, wl, collection = [])
spectra_high1 = plot_collection(spec_high1, wl, collection = [])
spectra_high2 = plot_collection(spec_high2, wl, collection = [])
# Read retrieved spectrum confidence regions
wl, spec_low2, spec_low1, spec_median, \
spec_high1, spec_high2 = read_retrieved_spectrum(planet_name, model_no_H2O['model_name'])
# Create composite spectra objects for plotting
spectra_median = plot_collection(spec_median, wl, collection = spectra_median)
spectra_low1 = plot_collection(spec_low1, wl, collection = spectra_low1)
spectra_low2 = plot_collection(spec_low2, wl, collection = spectra_low2)
spectra_high1 = plot_collection(spec_high1, wl, collection = spectra_high1)
spectra_high2 = plot_collection(spec_high2, wl, collection = spectra_high2)
# Produce figure
fig_spec = plot_spectra_retrieved(spectra_median, spectra_low2, spectra_low1,
spectra_high1, spectra_high2, planet_name,
data, R_to_bin = 100,
spectra_labels = ['Reference model', 'No H$_2$O'],
data_labels = ['STIS G430', 'STIS G750', 'WFC3 G141'],
data_colour_list = ['lime', 'cyan', 'black'],
plt_label = 'Effect of Removing H$_2$O')
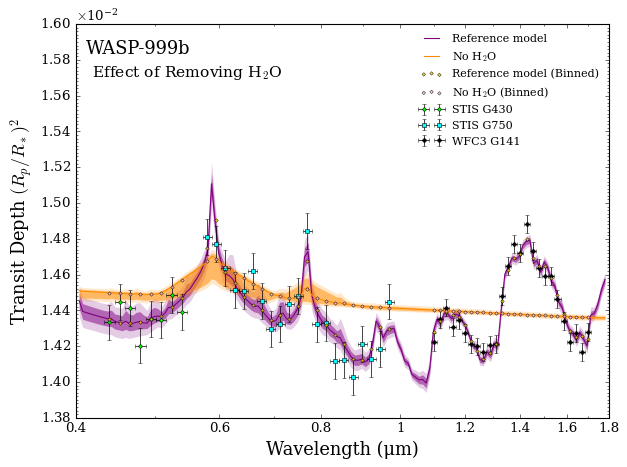
As you can see, even when the retrieval tries its best to minimise the residuals, without \(\rm{H}_2 \rm{O}\) there is simply no way to explain the data. This is the kind of scenario that results in a 23 σ detection.
What about \(\rm{TiO}\)?
[11]:
# Read retrieved spectrum confidence regions
wl, spec_low2, spec_low1, spec_median, \
spec_high1, spec_high2 = read_retrieved_spectrum(planet_name, model_ref['model_name'])
# Create composite spectra objects for plotting
spectra_median = plot_collection(spec_median, wl, collection = [])
spectra_low1 = plot_collection(spec_low1, wl, collection = [])
spectra_low2 = plot_collection(spec_low2, wl, collection = [])
spectra_high1 = plot_collection(spec_high1, wl, collection = [])
spectra_high2 = plot_collection(spec_high2, wl, collection = [])
# Read retrieved spectrum confidence regions
wl, spec_low2, spec_low1, spec_median, \
spec_high1, spec_high2 = read_retrieved_spectrum(planet_name, model_no_TiO['model_name'])
# Create composite spectra objects for plotting
spectra_median = plot_collection(spec_median, wl, collection = spectra_median)
spectra_low1 = plot_collection(spec_low1, wl, collection = spectra_low1)
spectra_low2 = plot_collection(spec_low2, wl, collection = spectra_low2)
spectra_high1 = plot_collection(spec_high1, wl, collection = spectra_high1)
spectra_high2 = plot_collection(spec_high2, wl, collection = spectra_high2)
# Produce figure
fig_spec = plot_spectra_retrieved(spectra_median, spectra_low2, spectra_low1,
spectra_high1, spectra_high2, planet_name,
data, R_to_bin = 100,
spectra_labels = ['Reference model', 'No TiO'],
data_labels = ['STIS G430', 'STIS G750', 'WFC3 G141'],
data_colour_list = ['lime', 'cyan', 'black'],
plt_label = 'Effect of Removing TiO')
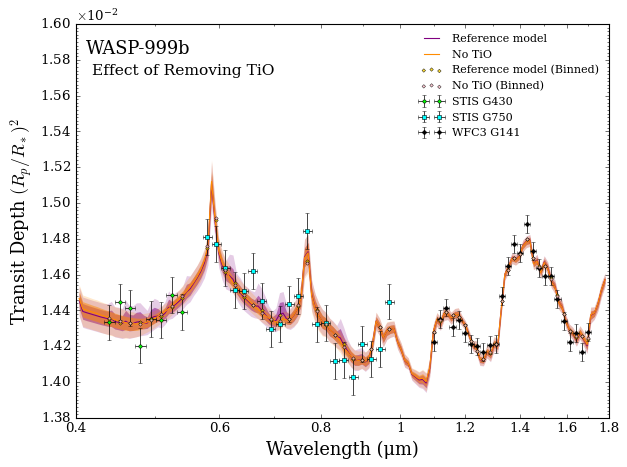
The retrieved spectrum looks almost the same, but with the possibility of \(\rm{TiO}\) bands no longer showing in the visible wavelengths. Since the overall quality of the fits are essentially the same, the Bayesian evidence favours the simpler model without \(\rm{TiO}\).